Glycinamide Ribotide Transformylase
Crystallization
14. Stura E.A., Johnson D.L., Inglese J., Smith J.M., Benkovic S.J., Wilson I.A. (1989) Preliminary Crystallographic Investigations of Glycinamide Ribonucleotide Tranformylase. Journal of Biological Chemistry 264:9703-9706.
Abstract: Crystals of glycinamide ribonucleotide transformylase have been grown from 0.4 to 1 M ammonium sulfate, 0.6 to 1 M sodium-potassium phosphate, or 0.65 to 1 M citrate in the pH range 4.5-7.0. The single crystals display variable morphology with varying pH. The crystals belong to the orthorhombic space group C222 with cell dimensions a = 141.4 Å, b = 98.2 Å, c = 103.5 Å. Co-crystals have also been obtained in the presence of the inhibitor 5,8-dideazofolate (KI = 18 mM) under similar crystallization variable morphology with varying pH. The crystals belong to the orthorhombic space group C222 with cell dimensions a = 141.4 Å, b = 98.2 Å, c = 103.5 Å. Co-crystals have also been obtained in the presence of the inhibitor 5,8-dideazafolate (KI = 18 mM) under similar crystallization conditions. Crystals of a chemically modified enzyme, iodinated at Cys-21, were grown under similar conditions within the pH range 6.5-7.0. These crystals are isomorphous with the unmodified enzyme. Crystals suitable for high resolution (less than 2.5 Å) x-ray diffraction studies have been obtained for each of the above.
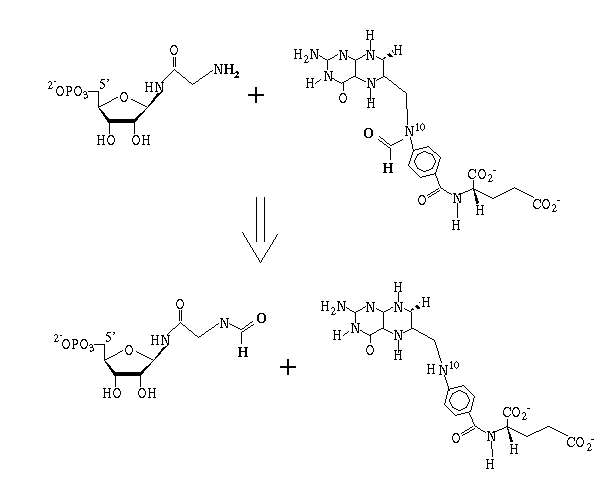
Reaction catalyzed:
Phosphate- "Native" Structure
29. Chen, P., Schulze-Gahmen U., Stura E.A., Benkovic S.J., Wilson I.A. (1992) Crystal Structure of Glycinamide Ribonucleotide Transformylase from Escherichia Coli. Journal of Molecular Biology. 227, 283-292.
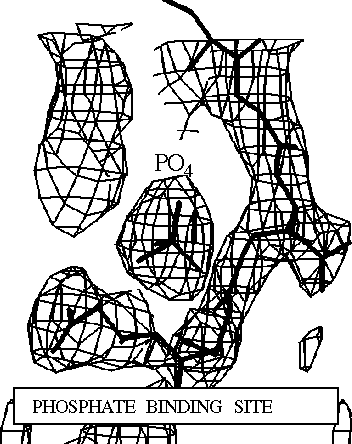
Abstract: The atomic structure of glycinamide ribonucleotide transformylase, an essential enzyme in purine biosynthesis, has been determined at 3.0 Å resolution. The last three C-terminal residues and a sequence stretch of 18 residues (residues 113 to 130) are not visible in the electron density map. The enzyme forms a dimer in the crystal structure. Each monomer is divided into two domains, which are connected by a central mainly parallel seven-stranded b-sheet. The N-terminal domain contains a Rossmann type mononucleotide fold with a phosphate ion bound to the C-terminal end of the first b-strand. A long narrow cleft stretches from the phosphate to a conserved aspartic acid, Asp144, which has been suggested as an active-site residue. The cleft is lined by a cluster of residues, which are conserved between bacterial, yeast, avian and human enzymes, and likely represents the binding pocket and active site of the enzyme. GAR-Tfase binds a reduced folate cofactor and glycinamide ribonucleotide for the catalysis of one of the initial steps in purine biosynthesis. Folate analogs and multi-substrate inhibitors of the enzyme have antineoplastic effects and the structure determination of the unliganded enzyme and enzyme-inhibitor complexes will aid the development of anti-cancer drugs.
Structure of complex with multisubstrate adduct
51. Klein C. Chen P. Arevalo J.H. Stura E.A. Marolewski A. Warren M.S. Benkovic S.J. Wilson I.A. (1995) Towards structure-based drug design: crystal structure of a multisubstrate adduct complex of glycinamide ribonucleotide transformylase at 1.96 Å resolution. Journal of Molecular Biology.[JMB Online Index] 249. 53-75.[Full Article]
Abstract: An inhibitor complex structure of glycinamide ribonucleotide transformylase (GAR-Tfase; EC 2.1.2.2) from Escherichia coli has been determined with a multisubstrate adduct BW1476U89 to an R-value of 19.1% at 1.96 Å resolution. The structure was determined by a combination of molecular and single isomorphous replacement using data from two different monoclinic crystal lattices and collecting data from crystals soaked in 20% (w/v) methyl-pentanediol as cryoprotectant for shock-freezing at -150°C. The multisubstrate adduct is bound in an extended crevice at the interface between the two functional domains of the enzyme. This inhibitor is positioned in the binding site by three sets of tight interactions with its phosphate, glutamate and pyrimidone ring moieties, while its intervening linker atoms are more flexible and adopt two distinct sets of conformations. The highly conserved Arg103, His108 and Gln170 residues that are key in ligand binding and catalysis (His108), have compensatory conformational variation that gives some clues as to their role in substrate specificity and in the formyl transfer. The molecular design of 1476U89 as a multisubstrate adduct inhibitor (Ki approximately 100 pM at pH 8.5), is confirmed as it closely mimics the shape, molecular interaction and combined binding constants of the natural 10-formyltetrahydrofolate (10-CHO-H4F; Km approximately 77.4 mM at pH 8.5) and glycinamide-ribonucleotide (GAR; Km approximately 8.1 mM at pH 8.5) substrates. The stereochemistry of this ligand complex suggests that His108 may act as an electrophile stabilizing the oxyanion of the tetrahedral intermediate that is formed as a result of the direct attack on the 10-CHO-H4F by the amino group of GAR. Structural comparison of the folate binding modes among GAR-Tfase, dihydrofolate reductase and thymidylate synthase reveals that folate derivatives bound to GAR-Tfase differentially adopt the trans conformation for the dihedral angle between atoms C-6 and C-9 providing a handle for targeting specific folate-dependent enzymes. The structural information derived from two different discrete conformations of the ligand in the binding site also suggests several leads for the de novo design of inhibitors of GAR-Tfase that may develop into useful chemotherapeutic agents.

