

Introduction
|
Either through luck, brilliance or blood, sweat and tears you have managed to grow crystals of your protein and even better, found they diffract AND there is a homologous structure in the PDB. This document is meant as a guide as to what to do next. It does not discuss the theory of crystallography (see references) and it is by no means complete. If you think it should include more material or fails at points then please let me know and I will try and fix it.  Russell Thom Russell Thom
Index of what is on this siteOther useful crystallography sitesMore information on the programs can be obtained by typing cdoc then more program_name.doc or http://www.dl.ac.uk/CCP/CCP4/dist/html/INDEX.html |

|
In processing you want to: Index the image correctly To do this we use one of a number of programs: XDS if you use the multiwire seimens detector MOSFILM |

|
This is a program to process data from the Siemens multiwire collector. For information about this program look up xds.doc by typing cdoc then more xds.doc XDS works via a number of steps ; XYCORR-calculates spatial corrections. INIT-determines initial background, anode wire modulation correction factors, and the trusted region on the detector surface. COLSPOT-collects strong diffraction spots occurring on the first nFRAME data frames. IDXREF-finds crystal orientation from spot locations and refines all parameters. COLPROF-collects 3-dimensional reflection profiles. PROFIT-estimates reflection intensities from profiles. CORRECT-corrects intensities for decay, absorption and detector surface sensitivity. GLOREF-refines all diffraction parameters. In order for XDS to work a command file must be in place, this is known as XDS.DATA and looks something like this XDS
|
|
What it means Line no. 1. Program we want to run, type in XDS (all the programs) or part i.e. CORRECT.
5. (a) x-ray wavelength (Angstrom). 6. (a) The number of the first data frame collected. 7. (a) Distance between crystal and detector (mm). (b) Unit cell parameters a, b, c (Angstrom) and alpha, beta, gamma (degrees). 9. (a) Number of minutes the program should wait before next
expected data frame-file will be available. 10. Integer array of 12 numbers providing a possibility of re-indexing the reflections in the final CORRECT step. Results of XDS can be seen by typing. |
|
Denzo has a nice printed manual termed HKL which describes a number of programs used in processing, it is well worth a read!!Also there are a few tutorials on the web. One of the best one can be found at the Department of Crystal and Structural Chemistry of the Bijvoet Center, Utrecht A brief introduction on how to get started is given below Open up two windows: window one- image windowwindow two- command window To begin processing using DENZO you need a param.den file(this is an ammalgamation of the site.dat and myexperiment.dat files in the manual) [For the BigDIP:] format dip 2030b |
|
The idea in processing is to try and index your intensities and determine a unit cell for your data.Using the param.den file we begin to specify the information we know best (wavelength, crystal to detector distance, detector type)and from this try and calculate information which we do not know:- unit cell. The other parts of the command file are variables in the experiment due to equipment and errors and during processing we are trying to determine these accurately. To run param.den setup denzo denzo (once you have setup denzo don't keep doing it as it will fill up your path length, type echo $PATH to see this) when the program appears to have stopped type @param.den @auto.den use partials position [default] peak search file 'peaks.file'write predictions fit cell crystal rotx roty rotz x beam y beam print zones go The program will give you an X and Y chi squared value followed by a list of errors. Using the Keywords to alter the parameters you want to try and get your chi squares close to one for your first image. Corrections may have to made to the box sizes in which the peaks are looked for and this is done by altering the lines spot, background and box in the parameter file. By looking at the image window it can be seen if the box is covering the spot correctly (example shown in figure 1). Figure 1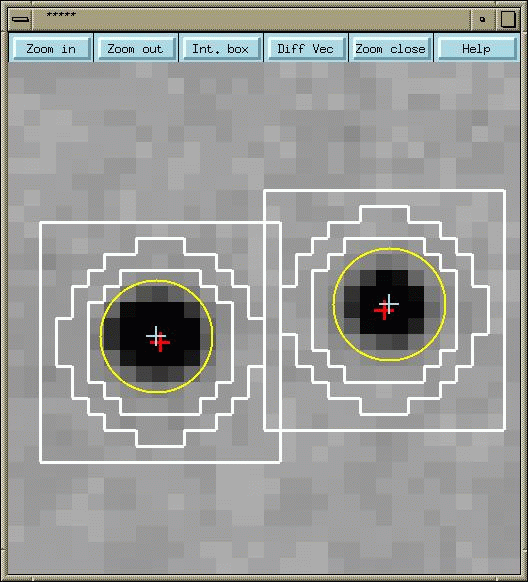 If some of the spots are not covered by the predictions you should try altering your mosaicity. Having autoindexed the data for your first image, the information you have gained (refined experimetal variables, unit cell, etc should be input back into the param.den) Using this new param.den and the file refine.den all the images can be autoindexed by subbing process.com. START REFINEMENT fit y beam x beamgo go go go go fit angular offset radial offset go go go go go fit crystal rotx roty rotz refine partiality go go go go go fit crossfire y x xy go go go go go fit cassette roty rotx go go go go go fit cell go go go go calculate go end of pack The "go" tells denzo how many times to refine that command Now process.com is used
|
|
setup denzo denzo << EOF@param.den @refine.den EOF Examine the images have been processed correctly by checking its .log file and .x file. The hkl manual gives good explanations of what different columns etc mean in .log and .x files. A little program devised by Paul setup paule denzolog process.log This will allow you to print nice graphs about various statistics of the data |

|
Scalepack is used when merging data together to ensure all the intensities are scaled correctly in regard to one another given slight changes in crystals or the way the data was collected. Like denzo it is part of the HKL suite and the documentation is quite good in the printed manual. To run scalepack setup denzo hkl matrix -1 0 0 0 0 -1 0 -1 0 format xds profit.hkl add 40 sector width 2.0 frame width 0.2 profiles summed FILE 3 '../PROFIT.HKL' eof-scale |
|
Information such as the space group and the unit cell size is gained from the previous processing packages such as xds or denzo. rotx, roty and mosxx are fitted to film and not to crystal unless the crystal is cryocooled. Again to check and see if scalepack has worked we use more scalepack.log the important bits are -Mosaicity which should not vary too much. Remember to put refined cell paramaters from post refinement with scalepack into scalepack.com before final run. This improves merging Rs lots. |
|
Scalepack2mtz |
Once scaling of the data has been completed the formatted reflection file has to be converted to mtz format for the following programs to accept the data. To do this a program called F2MTZ is used information information on this program can be gained from http://www.dl.ac.uk/CCP/CCP4/dist/html/f2mtz.htmlTo run this program a file called scalepack2mtz.com was formed scalepack2mtz \hklin gst2t.sca \ hklout GST1_2xtals.mtz \ << eof SYMM 4 END eof After conversion to mtz format (check make of .log file to check all has gone according to plan the data was ready to be enter into the next program truncate. |
|
TRUNCATE |
The program reads a file of averaged intensities from SCALEPACK2MTZ and writes a file containing mean amplitudes and the original intensities. It allows us to analyse the data to see if it shows sensible crystallographic behaviour. If anomalous data is present then F(+), F(-), with the anomalous difference, plus I(+) and I(-) are also written out. The amplitudes are put on an approximate absolute scale using the scale factor taken from a Wilson plot For information on this program look at http://www.dl.ac.uk/CCP/CCP4/dist/html/truncate.html The command file used to for Truncate looks like this truncate.com truncate \HKLIN GST_2xtals.mtz \ HKLOUT truncated.mtz \ << eof TITLE GST 2crystals 26/2/98 xds data and denzo LABOUT F=F SIGF=SIGF NRESIDUE 428 TRUNCATE no eof In this case the TRUNCATE no in the second last line means the program takes the square root of the intensities, setting any negative ones to zero. |
|---|
|
MTZ 2 VARIOUS |
This produces a reflection file written to HKLOUT in a suitable form for other systems such as MULTAN, SHELX, TNT,X-PLOR or any other Fortranic ASCII format, from an MTZ file on HKLIN. For information on this program look at http://www.dl.ac.uk/CCP/CCP4/dist/html/mtz2various.html command file used in this case is mtz2various \HKLOUT gst1.hkl \ hklin truncated.mtz \ << EOF LABIN FP=F SIGFP=SIGF OUTPUT SHELX RESOLUTION 100 3.0 END EOF This puts the data in a form AMoRe can read. i.e. ascii |
|
mtz 2 dump |
This dumps part or all of any mtz file as plain text. It gives header information about the range of observations etc. It can also convert the mtz fii.e. into ASCI format which can be read by AMoRe. Type mtzdump hklin filename.mtz then type go |

|
Jorge Navaza's state-of-the-art molecular replacement package. In molecular replacement you are trying to solve a structure of a molecule using a molecule homologous in structure. In MR a preliminary model of the crystal structure is obtained by first rotating then translating the model molecule in the crystal lattice. Once this has been accomplished we can calculate phases from the model and combine them with observed structure factor amplitudes. The structure factors thus obtained, and the corresponding electron density map, contain a strong bias toward the starting model, but are usually sufficiently close the allow refinement. AMoRe includes routines to run a complete molecular replacement. It reformats the data from the new crystal form, generates structure factors from the model, calculates the rotation function (tests agreement from the Patterson functions from the data and the model at various orientations), and the translation function (correlation between observed intensities and the cross vectors between the symmetry related molecules of the model as it is moved about the unit cell). The program then applies rigid body refinement to the solutions For information on this program look at http://www.dl.ac.uk/CCP/CCP4/dist/html/amore.html and also more aide-memoire To set this up in the directory you wish it to be type setup jorge SETA create hkl.d file create data.d file in i directory create dato.i3 To run csh ./e/job sub amore Go for coffee as this takes a while!!! Remember to look in the /o directory for the output, the .log file is for the experts i.e. Jorge. sort.s - describes resolution ranges collected and the reflections
used in AMoRe Tips for Molecular replacement Use the best start model available (obvious eh!!) Remove any atoms in the model that are known to differ from the target structure i.e. If side-chains are different to that from known sequence then clip them to ALA. Differences (insertions) in the loop regions can be removed, but be careful, including to many wrong atoms increases overall noise, but removing too many correct atoms reduces the signal you are looking for. Structure is normally conserved to a greater extent than sequence and two pieces of sequences may have different residues, but show a common fold. Low resolution data contains information mostly about crystal packing and the arrangement of solvent and so should be ommited from the search thus dont use data greater than 10Å. High resolution data will differ greatly between the homologous model and the target protein as this data describes the precise conformation of residues. Thus we want to use a resolution range of between 4-10Å If AMoRe is able to molecular replace your data then take the rotation and translation matrices from the mr2ic.s file in /o and put them in lsqkab.com . This reads in the model pdb and applies a rotation and translation and then outputs a new pdb file. #!/bin/csh -f WORKCD d/xyz1.d \ LSQOP testn1a.pdb \ DELTAS lsq_cat_residues.deltas \ << 'END-lsqkab' title ROTAT MATRIX 0.48986 -0.21954 -0.84371 -0.20192 0.91289 -0.35478 0.84810 0.34415 0.40286 TRAN 49.56 -41.85 -54.08 output XYZ fit WATOM 1 TO 10000 end 'END-lsqkab' If the pdb that your model is from contains hydrogens, these can be removed by running the program below to remove hydrogens |
|
# |

|
Pdb to map |
After this we can run sfall.com which calculates structure factors (Fcalc) and phases (Acalc) from our model. Our model isn't very good at this stage so we use SigmaA to calculate weights for each reflection (FOM) figures of merit. To try and improve the phases density modification is performed using the program DM. |
|
DM |
Density Modification Solvent flattening can be carried out because of a few truths found for all crystal strucutres. Solvent fills the gaps formed by imperfect packing of the molecules. This solvent is usually disordered from cell to cell and so in the diffraction structure the solvent electron density can be thought of as constant and zero. Also as proteins have fairly similar proportions of atomic types throughout the cell (C,N,O) according to the constraints of atomic spacing we can use this predictable atomic makeup to predict density values in the protein region (histogram matching). This is works even better if non crystallographic symmetry exists in the assymetric unit as averaging can be performed. Humble apologies, but I have not included this yet dmaverage is available if this is any help to you |
|
fft.com fast fourier transform |
This program is used to generate a map and extend it over our
model. Unlike visible wavelengths, x-rays cannot be focussed by a lens, the
fourier transform is a way the computer can act as a lens, using phases and
intensities to generate electron density. As each diffracted x-ray arrives to
the film it produces a reflection which can be described by a fourier series.
The fourier series that describes a diffracted ray is called its
structure-factor equation. This program converts these structure factors to
p(x,y,z) (electron density). A good place to learn about fourier transforms is
in the book Crystallography made Crystal Clear .
#!/bin/csh -f If you are not averaging your map then the file .ext can be put straight into mappage if you are going to alter your model in O, or it can be imported into Quanta. |
|
Mappage |
>Mappage setup o When it is finished it will say toodle pip and you now have a map file to put in to your o macro. Go use O or Quanta and build your structure. |
|
Judging Electron density |
When you have drawn your map in O or quanta then you should be able to see a large difference between solvent and protein. If the map is contoured at 1 sigma the protein map should have connected regions clearly separated from the solvent and these regions should all be the roughly the same height. Look out for model bias if using molecular replacement. This is seen as parts of the map having too much or too little density, with respect to the residue at that site. Model bias occurs, because the phase portion of the fourier transform makes up a large part of the synthesis, resulting in the homologous model being seen, rather than the target protein. Aromatic residues present in the target, but not in the model should show through in the map. Different types of maps can be used to try and overcome phase bias. OMIT maps - In this case residues with little electron density are removed and we rely on the fourier transform principle that every point in real space is contributed to by every point in reciprocal space. This means that the other atoms will contribute phases to give the correct image of the portion left out. DIFFERENCE maps - Fo-Fc maps.These will show unaccounted density and allows density contributed from the model and the target structure to be seen. |
|
Non -Crystallographic Symmetry By now you will know your proteins space group off by heart, but from looking at the crystal packing in SETOR earlier you might have noticed that you have more than one molecule in the asymmetric unit. These molecules may share NCS or local symmetry meaning that they show symmetry within the asymmetric unit. This can be used to produce averaged maps in which the noise will tend to cancel out and phases will be improved. |

|
ncsmask creaks a mask file which prevents the overlaps between the maps of the molecules being averaged. The matrices are specified so that the program can remove any overlaps. The pdb you read in should be a monomer from ncsmask xyzin solutiona.pdb mskout gst1a.msk << eof GRID 106 152 212 |
| Density modifiction |
As discussed earlier density modification of averaged maps is
a powerful technique to improve the phases of the target model.
dm HKLIN gst1mod1.mtz \ dm should have much better phases !! it will also have improved the matrices, so take these from the log file. As usual they are in a different format, so take care when putting them into mapprot! |
|
Calculating an average map Maprot |
MAPROT is used to generate an averaged map by transforming the electron density from molecules B, C and D onto A. maprot mapin gst1.ext mskin gst1a.msk wrkout gst1_av.map |

|
This will explain how to perform refinement from an averaged map, but if non averaged maps have been used input the pdb created form O or quanta directly into refmac and avoid Pdbset.com. Above lsqkab.com was used to calculate matrices from A to B, C and D, then using the averaged map only molecule A was built into. To reverse this process for refinement a pdb containing coordinates for molecules B, C and D must be created from molecule A . To do this pdbset.com is used. The matrices are those that were gained from lsqkab when the averaged map was built.
pdbset XYZIN muta_ref1.pdb \ This will generate a new pdb file and a log file. The pdb file can be examined using SETOR to check that the transform looks correct. |
|
Refmac
|
This is not the only refinement program available, but it is the only one I have so far used. Xplor another refinement program uses energy minimisation to refine data. Its manual can be found here, but I have not used this yet so sorry no more help than that at the moment. The refmac program minimizes the coordinate parameters to satisfy either a maximum likelihood or Least Squares residual. Geometric constraints can be used by including the program Protin which analyses the protein geometry and produces an output file which contains restraints for Refmac. Refmac also produces an HKL output file containing weighted coefficients for SigmaA, weighted difference FoFc and 2FoFc maps. #!/bin/csh -f eof |
|
[included only when water is added] Refmac will produce a log file, look for decreasing R factor and more importantly decreasing free R correlation factor should increase look at distances to see how many it is saying are wrong this should decrease with each cycle eventually convergence will occur and R free etc will not improve further. A new mtz file and pdb file is created in the scratch space of the computer refmac was run on. Use these files in fft.com to generate new maps. Type something like cp $CCP4_SCR/GST1_6.mtz . in the directory you wish to copy to, in the window of the computer you ran refinement in. Refmac is able to calculate new weighted phase and observation coefficients, fft.com should be told to look for these. |
|
fft.com -following refinement This map is for all the molecule, but as before it would be helpful to work on an averaged map, Lsqkab_averageing.com is used to generate rotation and translational matrices as before. These functions can then be put into ncsmask followed by maprot. Refinement is powerful and it can move your model quite a long way. Your new map should have better phases allowing easier building and the map should tell you where fairly biggish changes need to be made, |
|
fft_diff.com |
A difference map can be calculated to determine which areas of the density are being contributed to by the observed data and which from the calculated data. #!/bin/csh -f The difference between this command file and fft.com are the LABI difference weights calculated in refinement. This .ext file can now be put into maprot along with the mask created by ncsmask as before to generate a map. In the case of a difference map it is a good idea to follow the convention that the positive density is coloured red and the -ve density should be coloured green. This means residues in the green should move and those in the red should stay. |
|
Sample O |
Positve density (+sigma) Negative density (-sigma) The actual map using LABI F1=FWT PHI=PHWT in fft.com A sample O macro is shown below. Each map must be a different map_obj map_file gstslo1c_5a.brk |