Click here to find out how to optimise dSNAP and PolySNAP graphics displays for publication.
Click here for an overview comparing dSNAP, PolySNAP 3 and PolySNAP M.
PolySNAP 3 and PolySNAP M
PolySNAP 3
and PolySNAP
M are software packages designed to match and analyse patterns
utilising their full profiles. This allows for
quick and accurate identification of samples, even when working with
data of low, quality or where preferred orientation effects are
significant.
 The software provides an easy to use
interface to several powerful and novel statistical methods to rank
patterns in order of their similarity to any selected sample, allowing
unknowns to be quickly identified. In quantitative mode, given a
mixture pattern and potential pure phase patterns, it can identify
which patterns are in the mixture, and quantify their proportions
quickly and easily using a non-Rietveld based approach.
The software provides an easy to use
interface to several powerful and novel statistical methods to rank
patterns in order of their similarity to any selected sample, allowing
unknowns to be quickly identified. In quantitative mode, given a
mixture pattern and potential pure phase patterns, it can identify
which patterns are in the mixture, and quantify their proportions
quickly and easily using a non-Rietveld based approach.
The
matching procedure can be automated for high-throughput analysis,
allowing for large datasets to be analysed in a single run. Highly
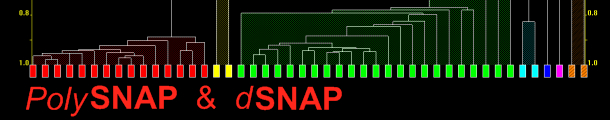
flexible graphical output is provided to summarise and visualise the
results. This highlights any unusual data, and means that time is not
wasted looking at the very many patterns that behave exactly as
expected.
PolySNAP M is designed to work with Powder X-Ray Diffraction patterns, and small datasets.
Its big brother, PolySNAP 3, can work on any one-dimensional data type - powder, Raman, DSC, IR, etc., as well as being able to utilise numeric data as well (e.g. melting points). These different techniques can be analysed individually, or much more powerfully, the results can be combined using an automatic weighting scheme to give an overall view of the dataset, which is invaluable to highlight any inconsistencies between different methods, as well as more accurate descriptions of what the samples are really like.
dSNAP
dSNAP is a free software
package to help users automatically classify and visualise the results
of database searches using the Cambridge Structural Database.
 With
the explosion in high quality structural determinations in the area of
small molecule crystallography, the problem of efficient mining of the
CSD is very relevant for structural chemists. The CSD represents an
enormously powerful resource, but faced with more than 300,000
structures, the attempt to extract meaningful chemical information can
be daunting. Many search algorithms and data mining strategies already
exist, but here we present a method of extracting chemical information
from such datasets using cluster analysis.
With
the explosion in high quality structural determinations in the area of
small molecule crystallography, the problem of efficient mining of the
CSD is very relevant for structural chemists. The CSD represents an
enormously powerful resource, but faced with more than 300,000
structures, the attempt to extract meaningful chemical information can
be daunting. Many search algorithms and data mining strategies already
exist, but here we present a method of extracting chemical information
from such datasets using cluster analysis.
By
exporting the complete geometric information for a fragment of interest
- all of the distances and angles - dSNAP provides facilities
for automatically classifying and visualising the results of
a search. It allows the user to assess those classifications
chemically, diminishing the need for tedious manual classification. The
real beauty of the method is in its interactivity and scalability - to
go from getting an overview of the entire dataset and easily spot any
outliers or errors, to the ability to ‘drill down’ within a cluster,
looking at more and more detailed differences between different
fragments.